Hto system: summary
Reference: Singari et al., Inducible protein traps with dominant phenotypes for functional analysis of the Drosophila genome.In this paper we presented a new transposon-based method, called Hostile takeover (Hto), that both expresses and tags endogenous proteins in Drosophila. Hto allows the investigator to express the target protein in a desired tissue under GAL4/UAS control. The target protein becomes tagged with a red fluorescent protein so it can be tracked in cells by fluorescence microscopy. A FLAG epitope tag is also added, allowing for detection on protein blots and purification from tissues.
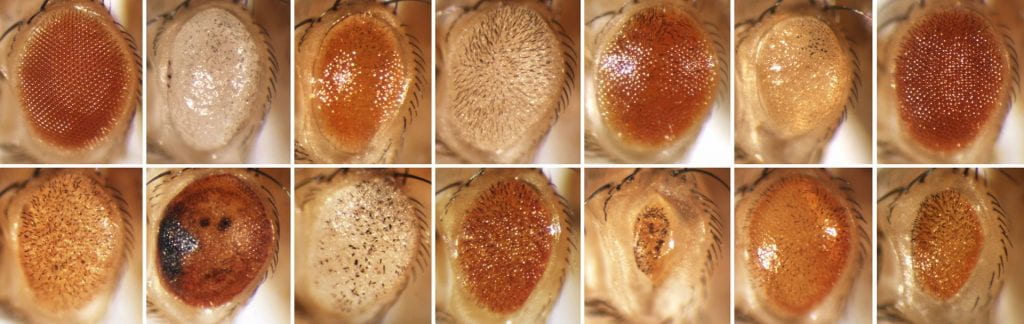
Using a simple genetic screen, we recovered several dozen Hto-bearing fly lines, of which 26 are described in Singari et al. Each one dominantly disrupts retinal development when expressed in the eye using GMR-GAL4. In several cases the Hto screen identified carboxy-terminal subfragments of proteins that can poison specific cellular processes; such fragments are rarely tested in conventional genetic screens. All of the target proteins so far recovered in this screen are conserved from flies to humans (or at least share conserved domains), and most are known regulators of cell biology or development. We are in the process of making the lines available for use, and the Hto system and individual lines are described in this site. A simple text list of the current set of target genes is at the bottom of this page.
The full size phenotype figures and other images can be found here.
Further info about Hto is here on the FAQ page.
Screening for new lines is described here.
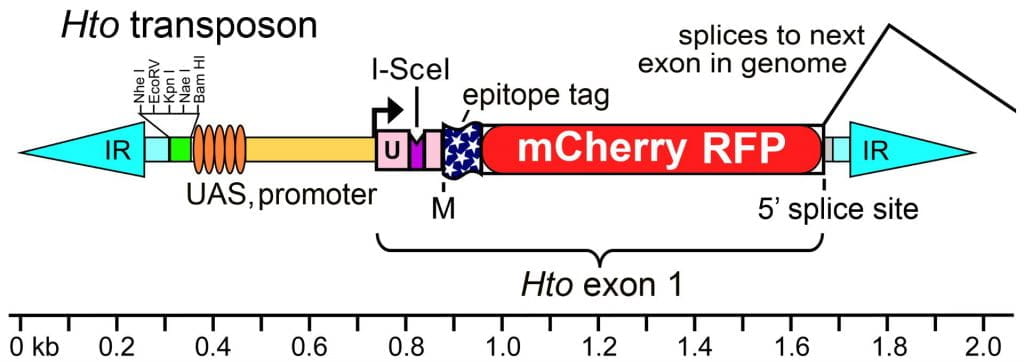
Above, map of the Hto transposon. Hto is flanked by inverted repeats (IR, cyan) from the Minos transposon. The internal portion of Hto carries a polylinker (green), the UAS (GAL4 binding sites) and basal promoter from the EP element, an artificial exon 1, and a canonical 5’ss. The exon 1 sequence includes a 5’UTR (U, pink) derived from a widely-expressed gene (sqh), with an added I-SceI endonuclease site (purple). This is followed by a start codon (M) and the coding region for the dual tag, 3xFLAG-mCherry red fluorescent protein (FLAG-RFP). Hto exon 1 may splice to the next downstream exon in the genome as indicated. See GenBank Acc. #JN049642 for the vector sequence.
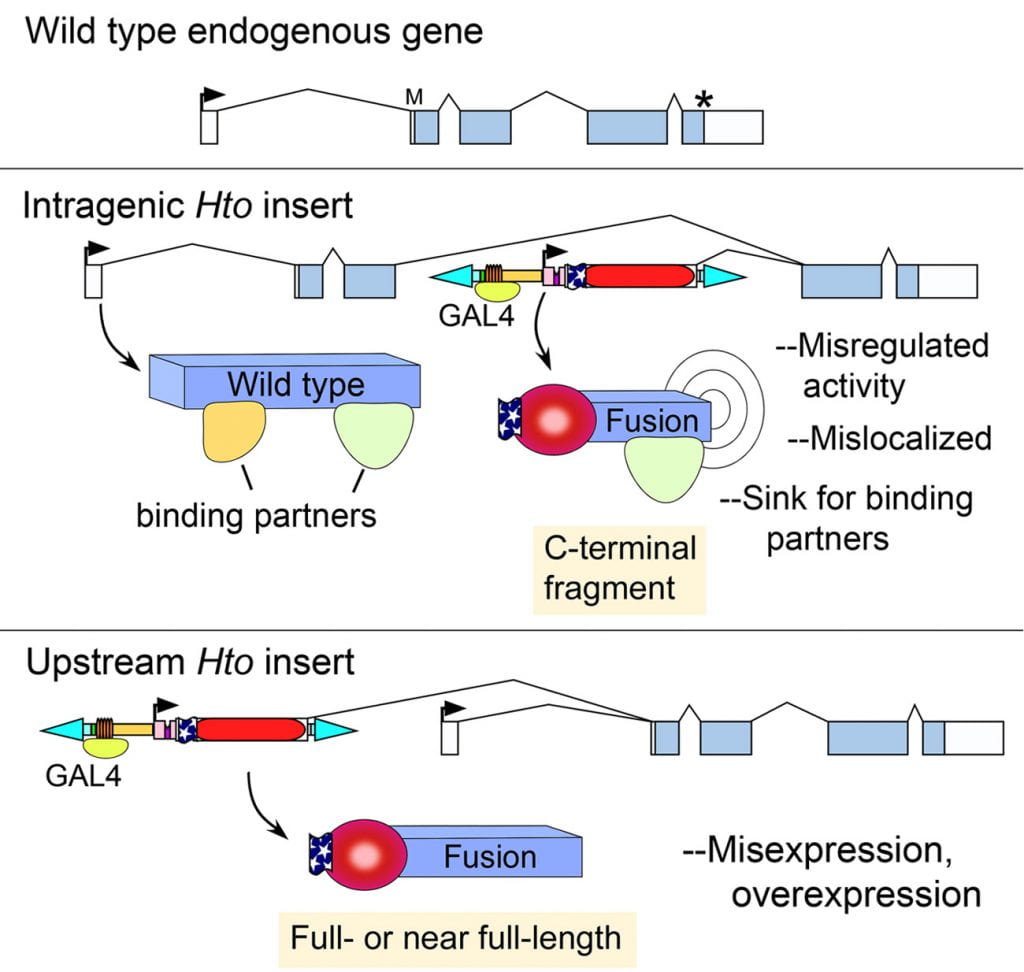
Above, Hto mechanism of action.
(Upper) Hypothetical wild type gene structure. Boxes, exons; arrow, transcription start; asterisk, stop codon.
(Middle) Intragenic insertion. When Hto inserts between the target gene’s start and stop codons, GAL4 induction of Hto can lead to expression of a tagged, C-terminal fragment of the endogenous target protein (“Fusion”). This may cause phenotypes by various mechanisms, as indicated (see Singari et al.)
(Lower) Upstream insertions of Hto will splice into exon 2 of the target gene. In the example here, this results in a tagged version of the full-length target protein, as long as there is an open reading frame from Hto exon 1 through to the target gene’s start codon.
Note that in both examples, the wild type gene products may still be produced using the target gene’s endogenous promoter, since Hto is minimally disruptive.
Here is a simple list of the genes that have been targeted by Hto insertions so far (see this page for phenotypes).
Published lines:
araucan (Iro-C)
bonus
CG11122
CG17181 / kahuli
CG34340
CG42235
cut
elbow B
iab-8, abd-A
klumpfuss
l(1)G0232
mamo / CG11071
mastermind
mir-274
nuclear fallout
pumilio
seizure
Shaven
Sox102F
Synaptotagmin 4
taranis
TfAP-2
vestigial
zfh2
Lines in prep:
Bantam
Cyclin E
Mir-184
pannier
scalloped