Hostile takeover Screening Method
This page explains how we isolated the published Hto lines and discusses the production of new lines.
For a more detailed discussion and cross schematic see Singari et al. 2014.
Here is the outline for the optimal method to get new inserts:
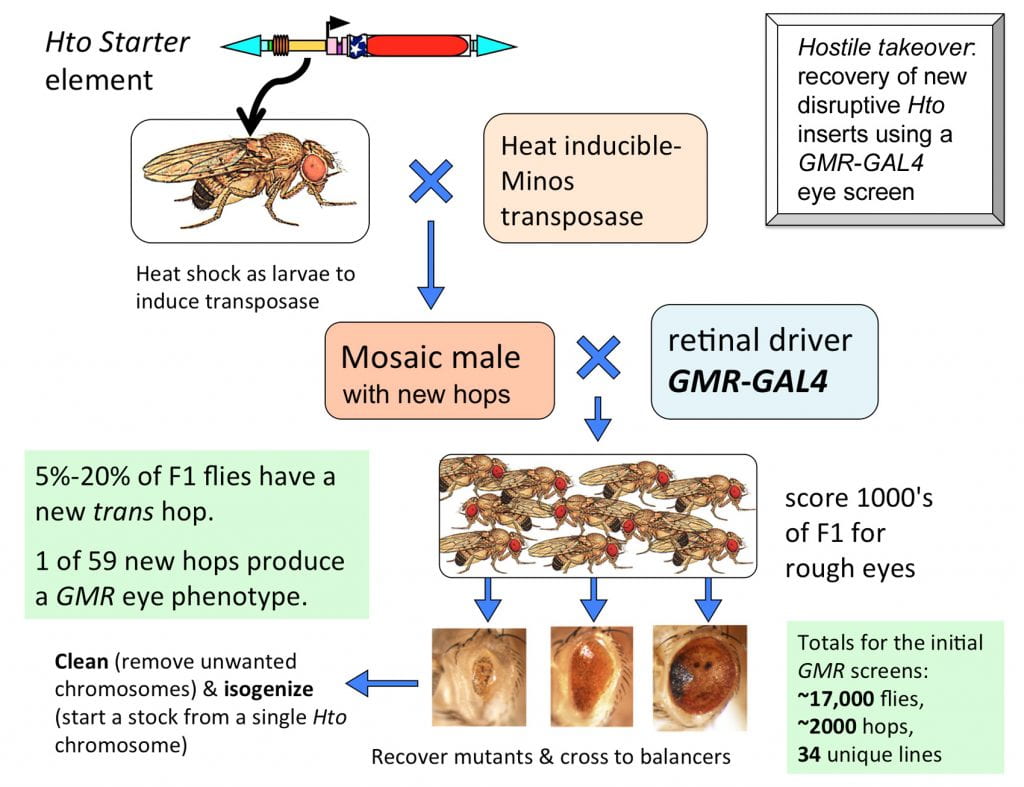
We constructed a Starter chromosome with two separate Hto inserts on the X chromosome. The Starter is crossed to the Minos transposase (MiT, on SM6,Cy) line. The larval offspring, Starter/Y; MiT/+; +/+, are heat shocked at 37 deg. C to generate new hops in the germ line. These mosaic males are crossed in small batches to a driver line such as GMR-GAL4. The F1 adults from this cross are screened for defects as they eclose, and crossed to balancer flies to make a stock.
Managing the crosses: We set up 2-3 mosaic males with a few GMR virgins in a vial, and brood it twice, to give 3 vials over ~7 days. It is important to give all of the vials from a given set of males a unique identifier (Set A/brood 1,2,3; Set B/brood 1,2,3, etc.) In this way, you can be certain that similar phenotypes from Sets A and B will be different inserts, while similar phenotypes from brood 1, 2 or 3 of Set A can be duplicates of the same insertion event. Be sure to keep the vials well fed and with optimal crowding; we usually split larvae to a new vial when they become overcrowded. This will help to ensure that less fit mutants survive to be screened. However, in practice, GMR-based mutants can eclose at any point in the brood and are not generally delayed in development.
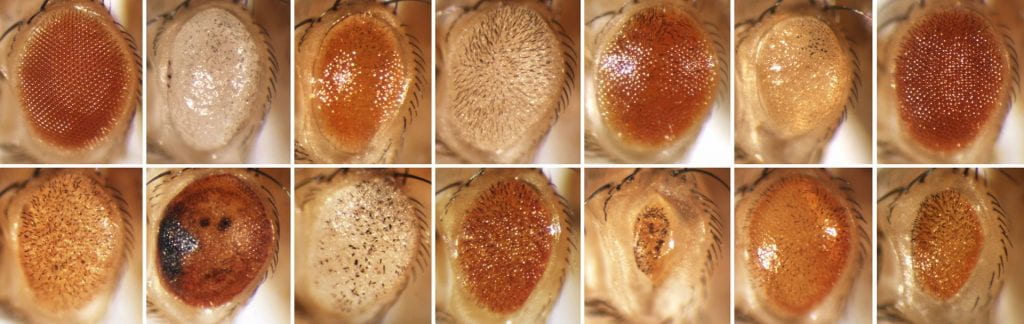
F1 Screening: You can expect about one mutant for every 3 standard vials screened; this is probably one per bottle if you do a bottle screen. We obtained ~1 strong mutant per 450 flies in our latest screen. With GMR, every mutant shows the phenotype in both eyes, so you can scan the flies without turning them over, making the actual screen very fast. (This is not true for eyeless-GAL4, which can often result in one normal and one malformed eye). If the mutant fly has inherited the Minos transposase chromosome, then the eyes will be a mosaic of mutant and wild type tissue (apparently the transposase transgene is expressed constitutively in the eye.) If the fly has mosaic defects in BOTH eyes, the insert is probably in the germline and will breed true after crossing out the transposase chromosome. If it has defects in only one eye, the insert is probably somatic and not in the germline, and the fly can be discarded.
Screens for expression and localization of the mCherry RPF tag: We have isolated numerous lines that do not have phenotypes, just based on their expression of RFP in the adult eye. These lines usually do NOT make protein fusions. Thus the mere expression of RFP is not an efficient way to get useful lines. Alternatively, one could express Hto in embryos or larvae and assess them manually for distinct RFP localization patterns. Preliminary results suggest this will be effective, but we have not yet done a full pilot screen. Based on our observations of somatic hops, there are scores of unique localization patterns that could obtained this way, independent of whether they cause misexpression phenotypes.